南京大学学报(自然科学版) ›› 2022, Vol. 58 ›› Issue (3): 369–376.doi: 10.13232/j.cnki.jnju.2022.03.001
• • 下一篇
基于深度卷积神经网络的RNA三维结构打分函数
- 1.南京大学物理学院,南京,210093
2.人工微结构科学与技术协同创新中心,南京,210093
An RNA tertiary structure scoring function based on deep neural network
Yuanyang Du1,2, Chengwei Deng1,2, Jian Zhang1,2( )
)
- 1.School of Physics, Nanjing University, Nanjing, 210093, China
2.Collaborative Innovation Center of Advanced Microstructures, Nanjing, 210093, China
摘要:
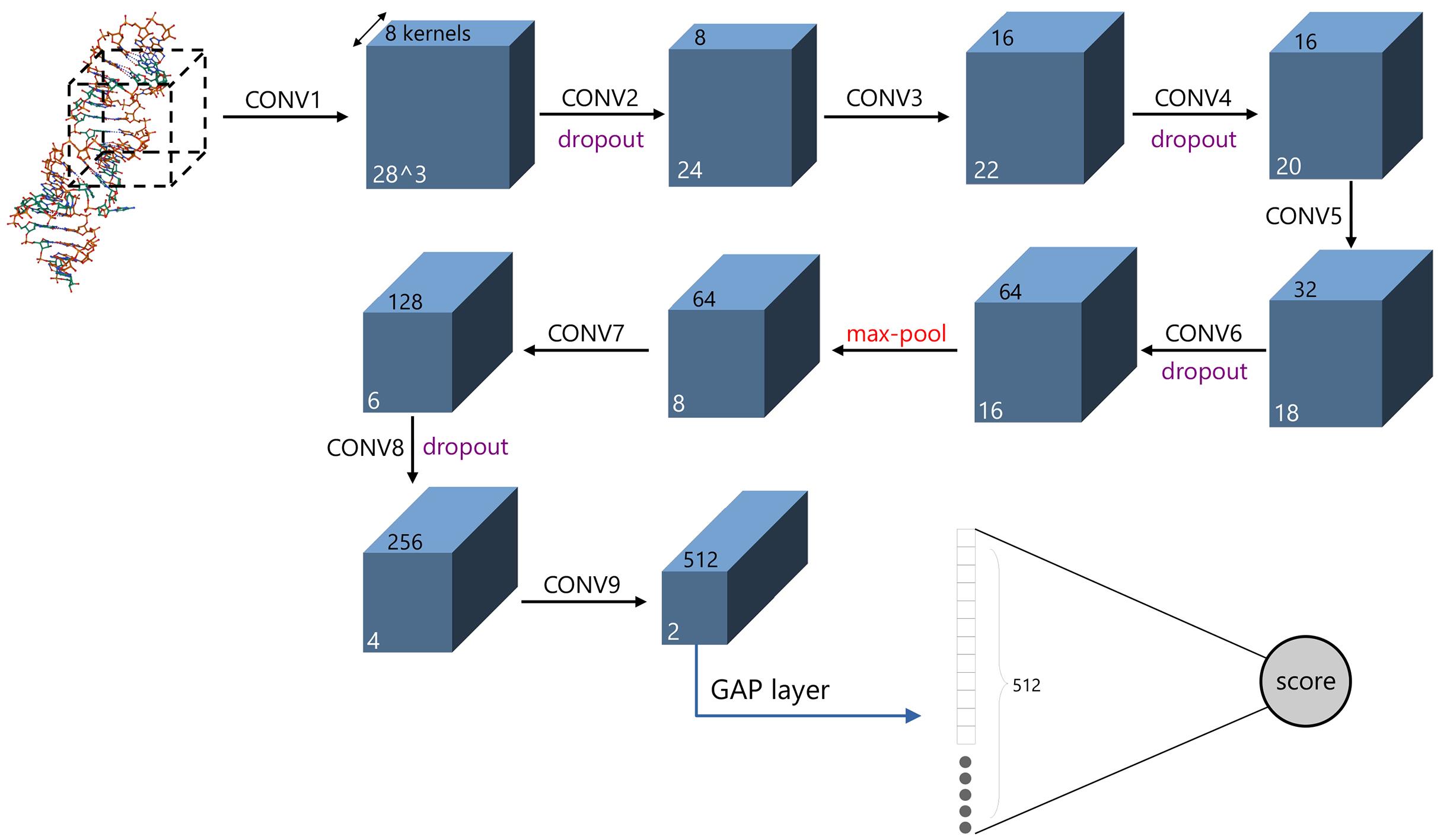
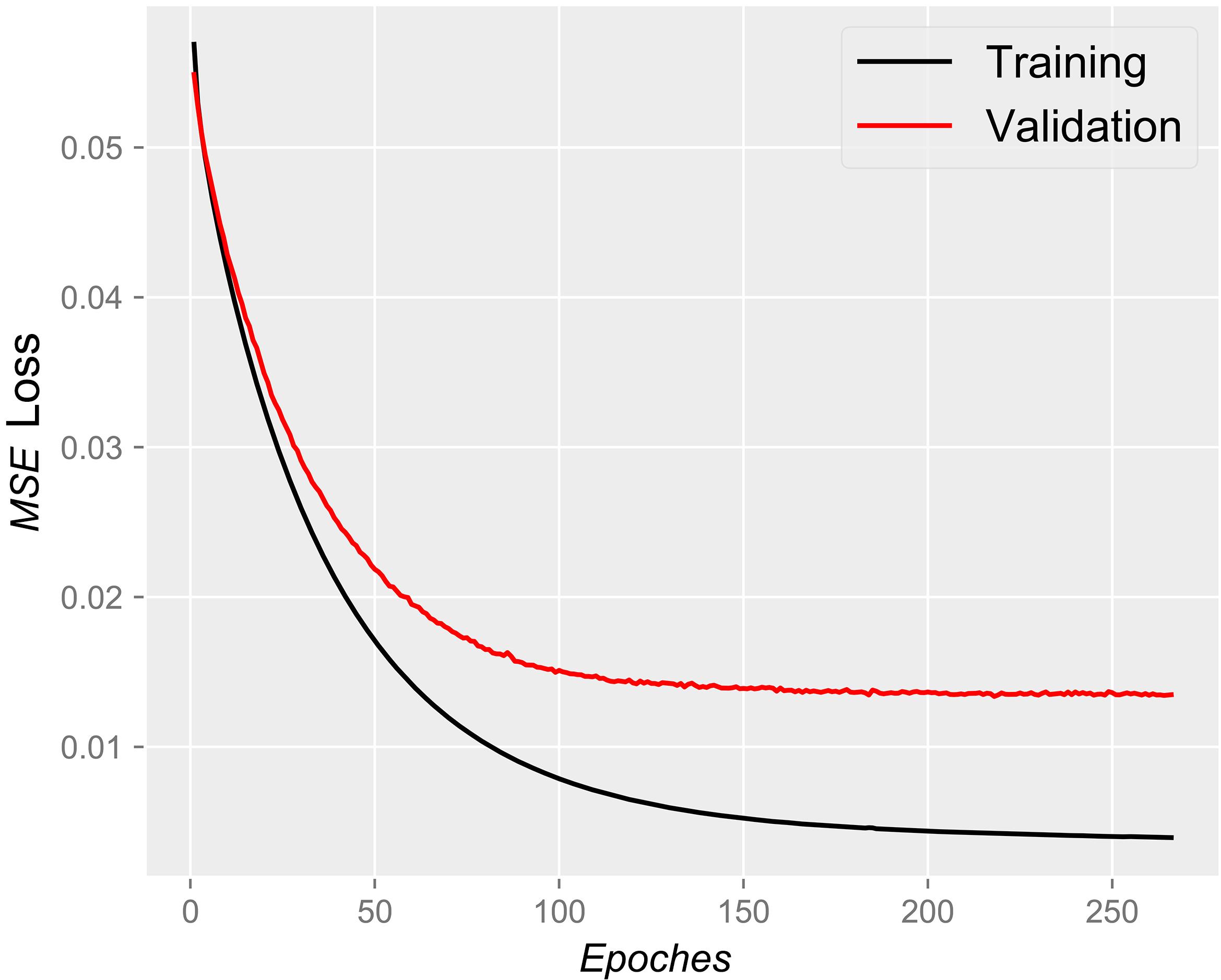
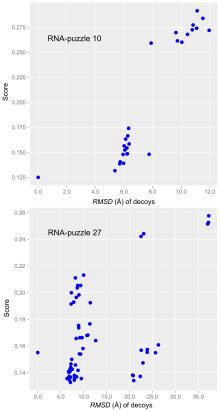
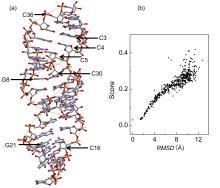
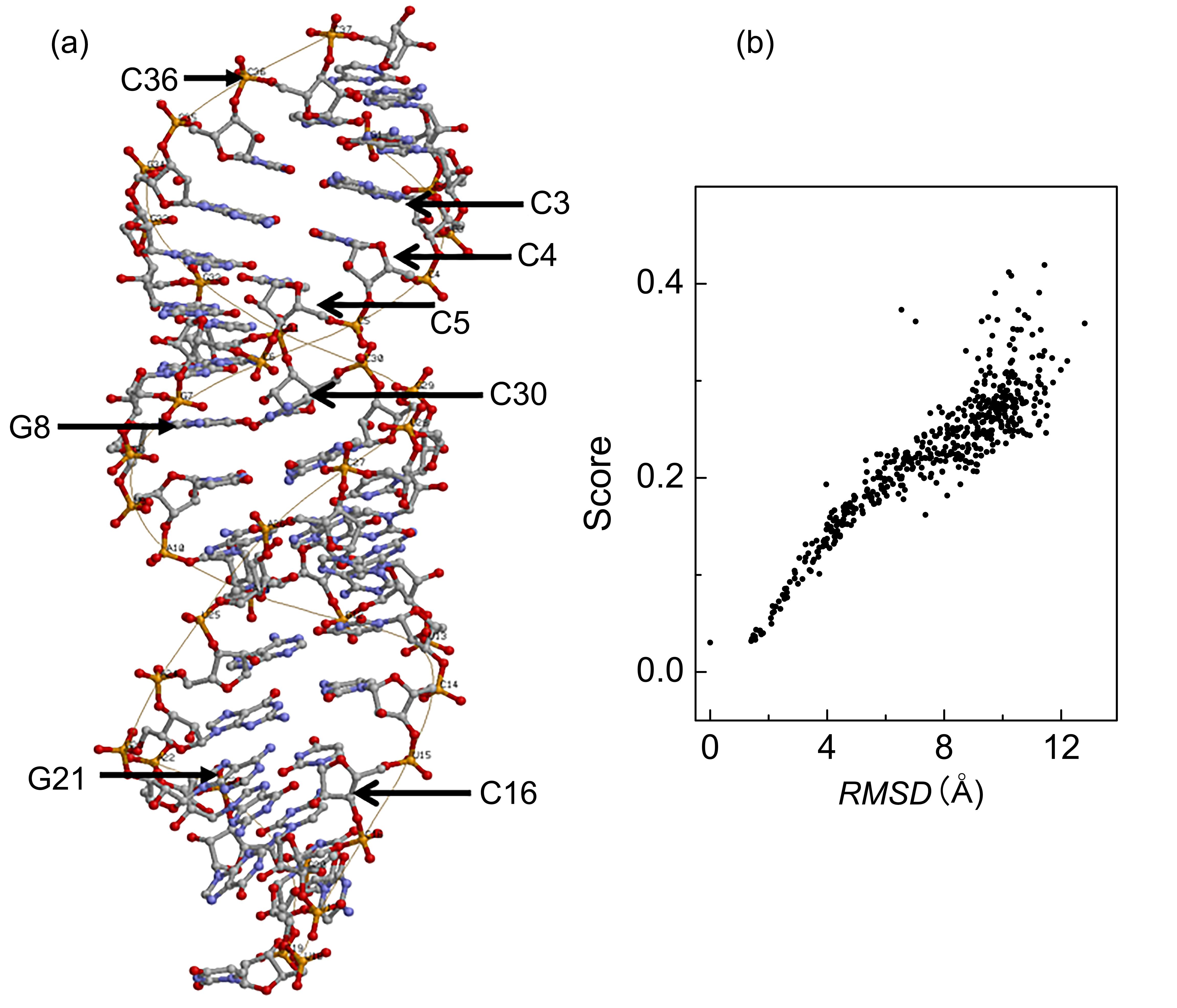
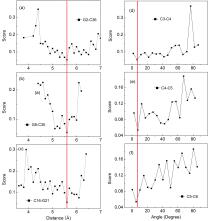
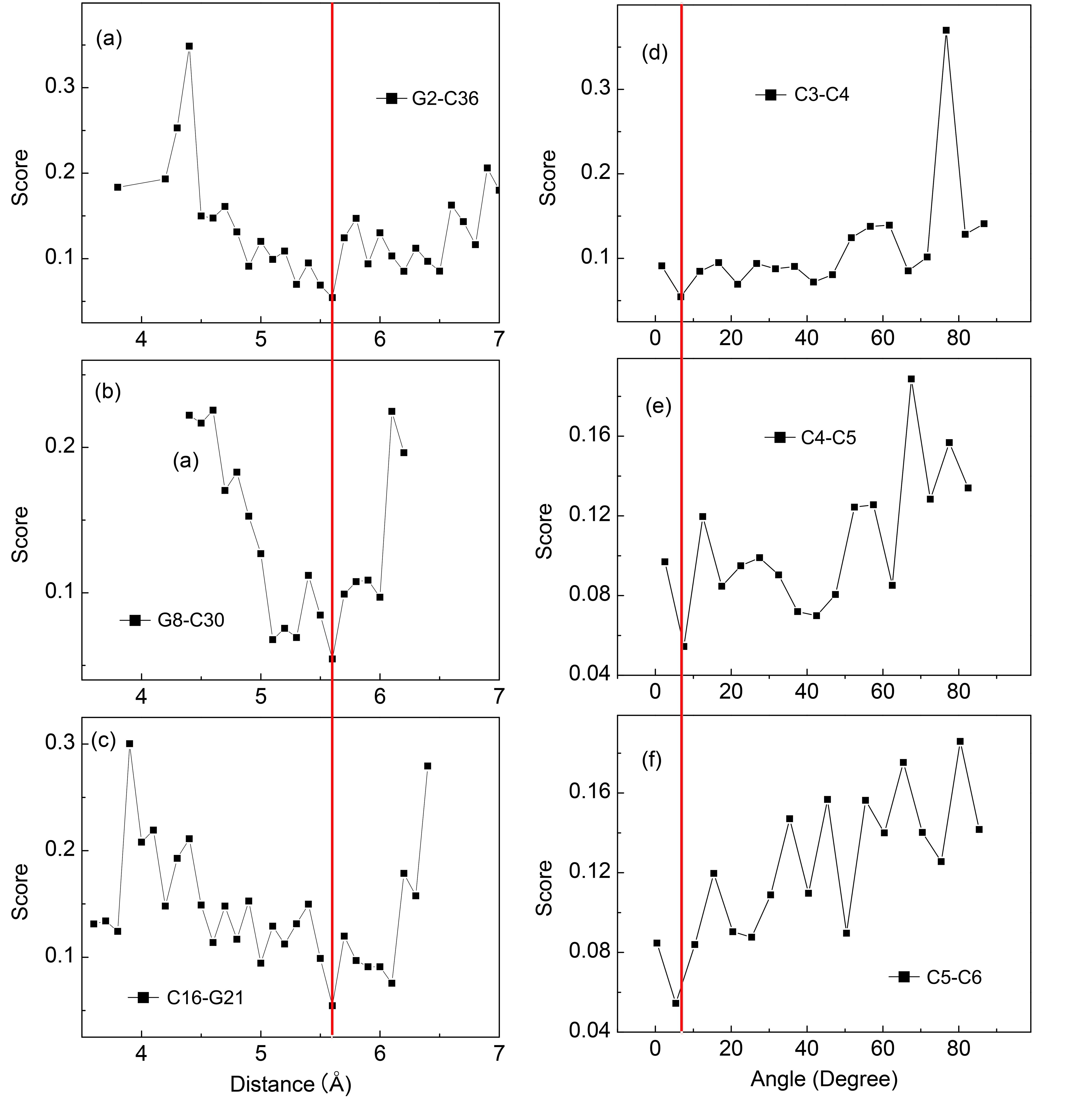
非编码RNA的三维结构对于人们理解和干预其生物功能具有重要的意义,从计算的角度发展RNA结构预测方法可以加速结构获取过程,对三维结构进行评分是进行结构预测的关键步骤.近年来,基于机器学习的方法,如AlphaFold2,已在分子结构预测领域取得了革命性的进展.基于深度卷积神经网络,建立了一个对RNA三维结构进行评估的方法.为了训练这一网络,建立了一个非冗余的含有422个RNA以及126600个decoys结构的数据集.训练得到的模型在RNA?Puzzles数据集上进行了测试,结果表明,在28个RNA中,网络从众多decoys中挑选出实验结构的正确率约为71.4%,这一结果比之前有所提高.另外,还对网络的工作机制进行了分析,发现神经网络对结构评分的倾向性和已知的物理化学知识相一致.
中图分类号:
- Q615
| 1 | Mercer T R, Dinger M E, Mattick J S. Long non?coding RNAs:Insights into functions. Nature Reviews Genetics,2009,10(3):155-159. |
| 2 | Geisler S, Coller J. RNA in unexpected places:Long non?coding RNA functions in diverse cellular contexts. Nature Reviews Molecular Cell Biology,2013,14(11):699-712. |
| 3 | Cech T R, Steitz J A. The noncoding RNA revolution?trashing old rules to forge new ones. Cell,2014,157(1):77-94. |
| 4 | Chen S J. RNA folding:Conformational statistics,folding kinetics,and ion electrostatics. Annual Review of Biophysics,2008(37):197-214. |
| 5 | Sun L Z, Zhang D, Chen S J. Theory and modeling of RNA structure and interactions with metal ions and small molecules. Annual Review of Biophysics,2017(46):227-246. |
| 6 | ?poner J, Bussi G, Krepl M,et al. RNA structural dynamics as captured by molecular simulations:A comprehensive overview. Chemical Reviews,2018,118(8):4177-4338. |
| 7 | Dans P D, Gallego D, Balaceanu A,et al. Modeling,simulations,and bioinformatics at the service of RNA structure. Chem,2019,5(1):51-73. |
| 8 | Shi Y Z, Wu Y Y, Wang F H,et al. RNA structure prediction:Progress and perspective. Chinese Physics B,2014,23(7):078701. |
| 9 | Wang J, Zhao Y J, Zhu C Y,et al. 3dRNAscore:A distance and torsion angle dependent evaluation function of 3D RNA structures. Nucleic Acids Research,2015,43(10):e63. |
| 10 | Wang J, Mao K K, Zhao Y J,et al. Optimization of RNA 3D structure prediction using evolutionary restraints of nucleotide–nucleotide interactions from direct coupling analysis. Nucleic Acids Research,2017,45(11):6299-6309. |
| 11 | Zhao Y J, Huang Y Y, Gong Z,et al. Automated and fast building of three?dimensional RNA structures. Scientific Reports,2012(2):734. |
| 12 | Tan Y L, Feng C J, Jin L,et al. What is the best reference state for building statistical potentials in RNA 3D structure evaluation? RNA,2019,25(7):793-812. |
| 13 | Tan Z J, Chen S J. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. The Journal of Chemical Physics,2005,122(4):044903. |
| 14 | Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge,USA:MIT Press,2016,775. |
| 15 | Jumper J, Evans R, Pritzel A,et al. Highly accurate protein structure prediction with AlphaFold. Nature,2021,596(7873):583-589. |
| 16 | Tunyasuvunakool K, Adler J, Wu Z,et al. Highly accurate protein structure prediction for the human proteome. Nature,2021,596(7873):590-596. |
| 17 | Li J, Zhu W, Wang J,et al. RNA3DCNN:Local and global quality assessments of RNA 3D structures using 3D deep convolutional neural networks. PLoS Computational Biology,2018,14(11):e1006514. |
| 18 | Huang B, Du Y Y, Zhang S,et al. Computational prediction of RNA tertiary structures using machine learning methods. Chinese Physics B,2020,29(10):108704. |
| 19 | Zhou B L, Khosla A, Lapedriza A,et al. Learning deep features for discriminative localization∥Proceedings of 2016 IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas,NV,USA:IEEE,2016:2921-2929. |
| 20 | Nawrocki E P, Burge S W, Bateman A,et al. Rfam 12.0:Updates to the RNA families database. Nucleic Acids Research,2015,43(D1):D130-D137. |
| 21 | Kalvari I, Argasinska J, Quinones?Olvera N,et al. Rfam 13.0:Shifting to a genome?centric resource for non?coding RNA families. Nucleic Acids Research,2018,46(D1):D335-D342. |
| 22 | Abraham M J, Van Der Spoel D, Lindahl E,et al. GROMACS User Manual version 2018.4. ,2018. |
| 23 | Cruz J A, Blanchet M F, Boniecki M,et al. RNA?Puzzles:A CASP?like evaluation of RNA three?dimensional structure prediction. RNA,2012,18(4):610-625. |
| 24 | Miao Z C, Adamiak R W, Blanchet M F,et al. RNA?Puzzles Round Ⅱ:Assessment of RNA structure prediction programs applied to three large RNA structures. RNA,2015,21(6):1066-1084. |
| 25 | Miao Z C, Adamiak R W, Antczak M,et al. RNA?Puzzles Round III:3D RNA structure prediction of five riboswitches and one ribozyme. RNA,2017,23(5):655-672. |
| 26 | Tishchenko S, Nikulin A, Fomenkova N,et al. Detailed analysis of RNA?protein interactions within the ribosomal protein S8?rRNA complex from the archaeon Methanococcus jannaschii . Journal of Molecular Biology,2001,311(2):311-324. |
| [1] | 陈轶洲, 刘旭生, 孙林檀, 李文中, 方立兵, 陆桑璐. 基于图神经网络的社交网络影响力预测算法[J]. 南京大学学报(自然科学版), 2022, 58(3): 386-397. |
| [2] | 高菲, 杨柳, 李晖. 开放集识别研究综述[J]. 南京大学学报(自然科学版), 2022, 58(1): 115-134. |
| [3] | 张玮, 赵永虹, 邱桃荣. 基于注意力机制和深度学习的运动想象脑电信号分类方法[J]. 南京大学学报(自然科学版), 2022, 58(1): 29-37. |
| [4] | 樊炎, 匡绍龙, 许重宝, 孙立宁, 张虹淼. 一种同步提取运动想象信号时⁃频⁃空特征的卷积神经网络算法[J]. 南京大学学报(自然科学版), 2021, 57(6): 1064-1074. |
| [5] | 孟浩, 刘强. 基于FPGA的卷积神经网络训练加速器设计[J]. 南京大学学报(自然科学版), 2021, 57(6): 1075-1082. |
| [6] | 陈磊, 孙权森, 王凡海. 基于深度对抗网络和局部模糊探测的目标运动去模糊[J]. 南京大学学报(自然科学版), 2021, 57(5): 735-749. |
| [7] | 倪斌, 陆晓蕾, 童逸琦, 马涛, 曾志贤. 胶囊神经网络在期刊文本分类中的应用[J]. 南京大学学报(自然科学版), 2021, 57(5): 750-756. |
| [8] | 杨静, 赵文仓, 徐越, 冯旸赫, 黄金才. 一种基于少样本数据的在线主动学习与分类方法[J]. 南京大学学报(自然科学版), 2021, 57(5): 757-766. |
| [9] | 李苓玉, 刘治平. 基于机器学习的自发性早产生物标记物发现[J]. 南京大学学报(自然科学版), 2021, 57(5): 767-774. |
| [10] | 贾霄, 郭顺心, 赵红. 基于图像属性的零样本分类方法综述[J]. 南京大学学报(自然科学版), 2021, 57(4): 531-543. |
| [11] | 普志方, 陈秀宏. 基于卷积神经网络的细胞核图像分割算法[J]. 南京大学学报(自然科学版), 2021, 57(4): 566-574. |
| [12] | 段建设, 崔超然, 宋广乐, 马乐乐, 马玉玲, 尹义龙. 基于多尺度注意力融合的知识追踪方法[J]. 南京大学学报(自然科学版), 2021, 57(4): 591-598. |
| [13] | 颜志良, 丰智鹏, 刘丹, 王会青. 一种混合深度神经网络的赖氨酸乙酰化位点预测方法[J]. 南京大学学报(自然科学版), 2021, 57(4): 627-640. |
| [14] | 崔鹤, 刘昆, 瞿晓磊. 基于紫外⁃可见光谱和机器学习方法的溶解性有机质吸附预测模型研究[J]. 南京大学学报(自然科学版), 2021, 57(3): 356-363. |
| [15] | 方志文, 刘青山, 周峰. 基于像素⁃目标级共生关系学习的多标签航拍图像分类方法[J]. 南京大学学报(自然科学版), 2021, 57(2): 208-216. |
|
||