乳腺癌(Breast Cancer,BC)是全球发病率最高的肿瘤类型,也是女性肿瘤死亡的首要原因.2015年中国大约有303600例新发乳腺癌病例以及70400例乳腺癌死亡,预计到2030年中国乳腺癌的发病率和死亡率将继续增加[1].目前乳腺癌的治疗策略主要包括内分泌治疗、靶向治疗和化疗,雌激素受体(ER)、孕激素受体(PR)、人表皮生长因子激素受体(HER⁃2)和Ki⁃67等是BC患者预测疗效的重要临床分子生物标志物.然而肿瘤的异质性和耐药性的出现使疗效显著降低,相同分子类型的BC预后也有差异,所以迫切需要确定新的治疗靶点和特定的生物标志物以提高BC患者的治疗疗效和预测预后的准确性.
高迁移率族框(High Mobility Group Box,HMGB)家族主要包括HMGB1,HMGB2和HMGB3,它们可以调节DNA复制、转录、重组和修复.HMGB家族基因在多种肿瘤中异常表达并发挥重要作用,有望成为肿瘤治疗的新靶点.其中,HMGB1和HMGB2在膀胱癌[2]、皮肤鳞状细胞癌[3]中过表达并与肿瘤进展相关,HMGB3在食管癌[4]、膀胱癌[5]和肝癌[6]中过表达并提示不良预后.实验研究表明HMGB1过表达能增强乳腺癌血管生成[7],HMGB1激活的成纤维细胞通过增加有氧糖酵解促进乳腺癌细胞转移[8].HMGB2在乳腺癌中表达高于正常乳腺组织,HMGB2表达是乳腺癌根治性切除术后的独立预后因素[9].但是目前HMGB3在乳腺癌中的表达情况、预后价值仍不明确,功能研究较少.此外,已经证实肿瘤浸润淋巴细胞(Tumor Infiltrating Lymphocytes,TIL)与多种肿瘤的预后相关,在免疫治疗中增强TIL水平在杀死肿瘤细胞方面具有非常显著的作用[10],而不同种类TIL与乳腺癌预后的关系仍不明确.
癌症基因组图谱 (The Cancer Genome Atlas,TCGA)等大型数据库中存储了大量的癌症患者的临床信息及基因组、转录组、蛋白组等各个组学数据,利用各种在线分析工具可以对这些数据进行交互探索与分析.因此,本研究利用多种生物信息学方法进行数据库挖掘,探究HMGB3基因在乳腺癌中的表达、预后和功能.此外,还分析了不同TIL水平对乳腺癌预后的影响,并探究HMGB3与TIL影响乳腺癌预后的可能机制,旨在为临床提供有助于改进治疗设计和预测BC患者预后的潜在生物标志物.
1 资料与方法
1.1 TIMER数据库泛癌表达及免疫预后分析
TIMER (Tumor Immune Estimation Resource)数据库是一个基于TCGA的数据分析平台 (https:∥cistrome.shinyapps.io/timer/)[11],通过TIMER的Diff Exp模块分析HMGB3在不同癌症中的表达情况,通过Survival模块计算不同免疫细胞浸润水平对肿瘤患者的预后影响.设置条件:
(1)Cancer Type:BRCA(Breast Invasive Carcinoma).
(2)Immune Inflitrates:B cell,CD8+T cell,CD4+T cell,Macrophage,Neutrophil,Dentritic Cell.
(3)Kaplan⁃Meier Curve Parameters:Split Percentage of Patients 50%,Survival Time Between (Month) 120.
1.2 GEPIA 和UALCAN表达分析
基因表达谱数据动态分析 (Gene Expression Profiling Interactive Analysis,GEPIA,http:∥gepia.cancer⁃pku.cn)[12]数据库是一个基于TCGA和GTEx数据的网页分析工具,可提供差异表达分析、轮廓图绘制、患者生存分析、相关基因分析等功能.使用GEPIA分析乳腺癌和正常组织之间的表达差异.筛选条件:
(1)Gene:HMGB3.
(2)Datasets Selection:BRCA,其他筛选框保持默认值.
UALCAN(http:∥ualcan.path.uab.edu/)[13]使用来自临床蛋白质组学肿瘤分析协会(Clinical Proteomic Tumor Analysis Consortium,CPTAC)证实或发现的数据集数据分析不同肿瘤蛋白质表达,可以通过UALCAN的“CPTAC analysis”功能探究HMGB3在乳腺癌和正常乳腺组织中的蛋白表达差异,利用UALCAN分析基于TCGA数据的乳腺癌的HMGB3启动子甲基化水平.
1.3 PrognoScan和Kaplan⁃Meier Plotter预后分析
PrognoScan数据库(http:∥dna00.bio.kyutech.ac.jp/PrognoScan/index.html)[14]基于大量公共可得的癌症微阵列数据集分析其中特定基因表达与癌症患者预后之间的关系,例如总体生存期(Overall Survival,OS)、无病生存期(Disease Free Survival,DFS)、无远处转移生存期(Distant Metastasis Free Survival,DMFS)、无复发生存期(Relapse Free Survival,RFS)和疾病特异性生存期(Disease Specific Survival,DSS)等.本研究通过PrognoScan数据库检索HMGB3不同表达对乳腺癌患者预后的影响.
Kaplan⁃Meier Plotter(http:∥kmplot.com/analysis/)[15]是一种基于生物标志物评估的在线荟萃分析工具,能基于大型样本数据集来评估不同生物标志物的表达对不同类型癌症患者生存的影响.通过Kaplan⁃Meier Plotter数据库分析HMGB3表达在不同临床因素,包括ER、PR、HER2、分子分型、淋巴结转移、肿瘤分级和TP53突变状态下与乳腺癌患者的无复发生存期RFS和无远处转移生存期DMFS预后的关系.设置条件:
(1)Cancer:Breast Cancer.
(2)Gene:HMGB3.
(3)Split patients by:median.
(4)Survival:RFS或DMFS.
(5)Follow up threshold:all.
(6)Probe set options:only JetSet best probe set.
通过Kaplan⁃Meier Plotter数据库中的泛癌数据集进一步筛选不同免疫细胞浸润水平下的HMGB3表达对乳腺癌患者总生存的影响.筛选条件:
(1)Gene symbol:HMGB3.
(2)Split patients by:median.
(3)Survival:OS.
(4)Follow up threshold:all.
(5)Cancer:Breast cancer (n=1090).
1.4 Coexpedia和STRING数据库功能分析
Coexpedia(https:∥www.coexpedia.org/)通过功能关联评估目的基因的共表达[16],分析HMGB3在乳腺癌中的分子调控网络.筛选条件:
(1)Human gene:HMGB3.
(2)MeSH incl.Neoplasms:Breast Neoplasms.
STRING数据库(https:∥string⁃db.org/)通过整合大量生物信息数据构建蛋白质⁃蛋白质互作网络并预测目标蛋白的生物学功能[17].通过基因本体论(Gene Ontology,GO)和京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)来进行目标基因的功能注释分析,显示细胞组成(Cellular Component,CC)、分子功能(Molecular Function,MF)、生物学过程(Biological Process,BP)和KEGG通路(KEGG Pathway)结果.筛选条件:
(1)List of name:输入HMGB3和通过Coexpedia获得的18个基因蛋白名称.
(2)Organism:Homo Sapiens.
(3)FDR<0.05.
1.5 统计学方法
采用数据库默认的统计学分析方法.在GEPIA和UALCAN中,对正常组织与乳腺癌组织的差异分析采用独立样本t检验.在PrognoScan中,使用单变量Cox回归模型来计算HR和P,应用R软件(版本4.0.4)将预后结果可视化.在对Kaplan⁃Meier Plotter的分析中,用Kaplan⁃Meier法绘制生存曲线,用log⁃rank检验来分析HMGB3表达与乳腺癌患者生存之间的相关性.所有数据以P<0.05视为有统计学意义.
2 结果
2.1 HMGB3在不同肿瘤中的表达情况
有研究报道HMGB3在多种癌症中过表达,本研究通过TIMER数据库分析得到了相似的结果.如图1a所示,HMGB3转录水平在多种肿瘤中高于正常组织,包括膀胱尿路上皮癌、乳腺癌、胆管癌、结肠腺癌、食道癌、头颈部鳞状细胞癌、嫌色肾细胞癌、肾乳头状细胞癌、肝细胞癌、肺腺癌、肺鳞状细胞癌、前列腺腺癌、直肠腺癌、胃腺癌和子宫内膜癌.但在肾透明细胞癌中,正常组织中HMGB3 转录水平高于肿瘤组织.通过GEPIA进一步验证了HMGB3在乳腺癌中表达高于正常乳腺组织(图1b).此外,通过UALCAN分析还可以发现,在蛋白表达水平上,乳腺癌中的HMGB3表达同样高于正常组织,如图1c所示.以上结果表明:乳腺癌中HMGB3转录水平和蛋白水平均高于正常组织,可能在乳腺癌的发生发展中发挥作用.
图1
图1
HMGB3表达水平及甲基化水平:(a) HMGB3基于TCGA数据分析的在不同类型肿瘤中的表达水平 (TIMER);(b) HMGB3基于TCGA和GTEx数据分析的在乳腺癌和正常乳腺组织中的表达 (GEPIA);(c)HMGB3蛋白在乳腺癌和正常乳腺组织中的表达 (UALCAN);(d)HMGB3启动子甲基化水平在乳腺癌和正常乳腺组织中的表达 (UALCAN)
CPTAC:Clinical Proteomic Tumor Analysis Consortium,临床蛋白质组学肿瘤分析联盟;TCGA:The Cancer Genome Atlas,癌症基因组图谱;
Fig.1
The expression level and methylation level of HMGB3:(a) HMGB3 expression in different types of tumors based on TCGA data analysis (TIMER),(b) HMGB3 expression in breast and normal breast tissues (GEPIA) based on TCGA and GTEx data analysis,(c) HMGB3 protein expression in breast and normal breast tissues (UALCAN),(d) expression level of HMGB3 promoter methylation in breast and normal breast tissues (UALCAN)
2.2 HMGB3的甲基化分析
2.3 HMGB3基因在不同乳腺癌数据集中的预后
为了研究HMGB3表达的升高是否影响乳腺癌患者的预后,通过PrognoScan数据库进一步探索HMGB3不同表达对乳腺癌患者预后的影响.图2给出了21个GSE数据集中的13个数据集,其HMGB3的表达水平与乳腺癌患者预后相关(P<0.05),其余数据集,其表达结果无显著统计学差异,在此略去.由图可见,GSE12276,GSE1378,GSE1379数据集的HMGB3高表达与患者较差的RFS相关;GSE9195数据集的HMGB3高表达与患者不良DMFS和RFS相关;GSE11121数据集HMGB3高表达与患者较差的RFS相关;GSE1456⁃GPL96,GSE1456⁃GPL97中的HMGB3高表达与患者不良DFS和RFS相关,GSE1456⁃GPL97中的HMGB3高表达还与患者不良OS相关;GSE7378,GSE4922⁃GPL96,GSE4922⁃GPL97数据集的HMGB3高表达提示患者较差的DFS;GSE3494⁃GPL96,GSE3494⁃GPL97数据集的HMGB3高表达提示患者DSS较差,但在E⁃TABM⁃158数据集中,HMGB3高表达提示患者有较好的DSS.以上结果都具有统计学显著差异(
图2
图2
PrognoScan数据库中不同数据集的HMGB3表达对乳腺癌患者预后影响
RFS:Relapse Free Survival,无复发生存期
DMFS:Distant Metastasis Free Survival,无远处转移生存期
DSS:Disease Specific Survival,疾病特异性生存期
DFS:Disease Free Survival,无病生存期
Fig.2
The prognostic effect of HMGB3 expression in breast cancer patients based on different datasets of PrognoScan database
2.4 HMGB3表达在不同临床病理因素下对乳腺癌预后的亚组分析
由于Kaplan⁃Meier Plotter数据库的样本量更大,可以通过Kaplan⁃Meier Plotter进一步验证HMGB3表达在乳腺癌患者中的预后作用.表1给出了不同临床病理因素下乳腺癌中HMGB3表达与临床预后的相关性,表中黑体字为P<0.05具有显著统计学差异的结果.由表可见,HMGB3与乳腺癌患者更差的无复发生存期RFS和无远处转移生存期DMFS有关(
表1 不同临床病理因素下乳腺癌中HMGB3表达与临床预后的相关性(Kaplan⁃Meier Plotter)
Table 1
| N | Hazard ratio | P | N | Hazard ratio | P | ||
| Total analysis | 2032 | 1.54 (1.32~1.29) | 2e~8 | 958 | 1.77 (1.35~2.32) | 3.1e-05 | |
| ER | ER positive | 902 | 1.49 (1.12~1.97) | 0.006 | 248 | 2.27 (1.03~5.02) | 0.037 |
| ER negative | 470 | 0.91 (0.67~1.24) | 0.55 | 270 | 1.21 (0.79~1.86) | 0.37 | |
| PR | PR positive | 511 | 1.41 (0.97~2.04) | 0.072 | 144 | 2.18 (0.74~6.38) | 0.15 |
| PR negative | 436 | 1.29 (0.92~1.81) | 0.14 | 266 | 1.38 (0.89~2.16) | 0.15 | |
| HER2 positive | 461 | 1.44 (1.06~1.95) | 0.019 | 260 | 1.55 (0.98~2.45) | 0.062 | |
| HER2 negative | 1571 | 1.48 (1.24~1.76) | 1.20E~05 | 698 | 1.41 (1.02~1.96) | 0.04 | |
Intrinsic subtype | basal | 417 | 1.16 (0.86~1.58) | 0.34 | 275 | 1.24 (0.78~1.99) | 0.36 |
| luminal A | 952 | 1.37 (1.07~1.75) | 0.011 | 361 | 1.56 (0.93~2.6) | 0.089 | |
| luminal B | 465 | 1.22 (0.9~1.64) | 0.2 | 203 | 1.24 (0.7~2.22) | 0.46 | |
| HER2+ | 198 | 1.7 (1.09~2.66) | 0.017 | 119 | 1.79 (0.97~3.3) | 0.059 | |
| Lymph node positive | 814 | 1.77 (1.39~2.27) | 3.90E~06 | 261 | 2.4 (1.45~3.97) | 0.00047 | |
| Lymph node negative | 574 | 1.06 (0.73~1.54) | 0.76 | 240 | 0.88 (0.44~1.76) | 0.72 | |
| grade | 1 | 113 | 0.97 (0.34~2.76) | 0.95 | 44 | NA | |
| 2 | 243 | 1.23 (0.75~2.01) | 0.42 | 91 | 1.93 (0.75~5) | 0.17 | |
| 3 | 481 | 0.91 (0.68~1.22) | 0.52 | 234 | 1.27 (0.76~2.12) | 0.37 | |
| TP53 | mutated | 132 | 2.04 (1.11~3.75) | 0.02 | 56 | 3.15 (0.98~10.14) | 0.043 |
| wild type | 82 | 0.42 (0.17~1) | 0.044 | 6 | NA | ||
2.5 肿瘤浸润淋巴细胞水平与乳腺癌患者的累积生存相关
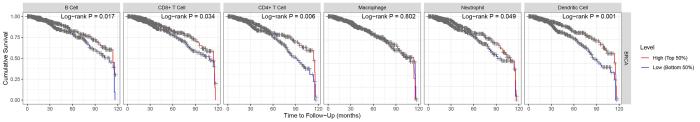
TIL独立预测癌症患者的总生存期和前哨淋巴结状态,已经有研究证实TIL与乳腺癌预后相关.在ER+/HER2⁃乳腺癌中,单因素分析表明TIL与患者无远处转移生存无关,但在接受化疗的患者中高水平的TIL与更好的无远处转移生存相关[21].而在三阴性乳腺癌中,高水平TIL可以提高患者的总生存期[22].然而这些研究都停留在总的TIL水平上,没有研究探讨不同种类TIL与乳腺癌预后的关系,这促使我们通过TIMER数据库研究在乳腺癌中不同TIL与乳腺癌预后的关系.如图3所示,高水平的B细胞、CD8+T细胞、CD4+T细胞、中性粒细胞和树突状细胞浸润的乳腺癌患者10年累积生存率更高,差异有统计学意义(所有
图3
图3
肿瘤浸润淋巴细胞水平对乳腺癌患者的10年累积生存的预后分析
Fig.3
The prognostic analysis of tumor infiltrating lymphocytes levels on the 10⁃year cumulative survival of breast cancer patients
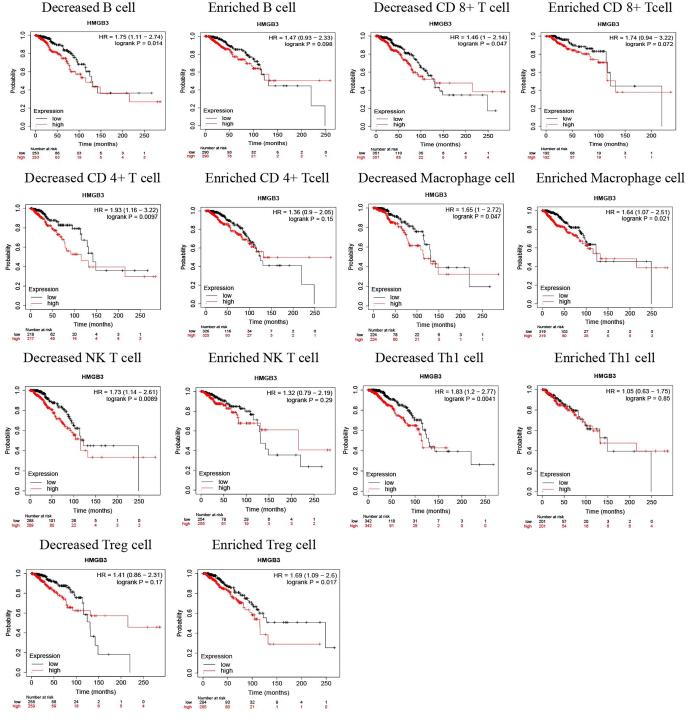
2.6 HMGB3表达在不同肿瘤浸润淋巴细胞水平下对乳腺癌患者的预后分析
已知HMGB3表达影响乳腺癌预后,TIL水平也与乳腺癌预后相关,通过Kaplan⁃Meier Plotter进一步探究TIL水平是否对HMGB3跟乳腺癌总生存率(Overall Survival,OS)的关系有影响.如图4所示,在低的B细胞(
图4
图4
不同肿瘤浸润淋巴细胞水平下乳腺癌HMGB3表达与总生存的相关性
Fig.4
Correlation of HMGB3 expression and overall survival in breast cancer with different tumor infiltrating lymphocytes levels
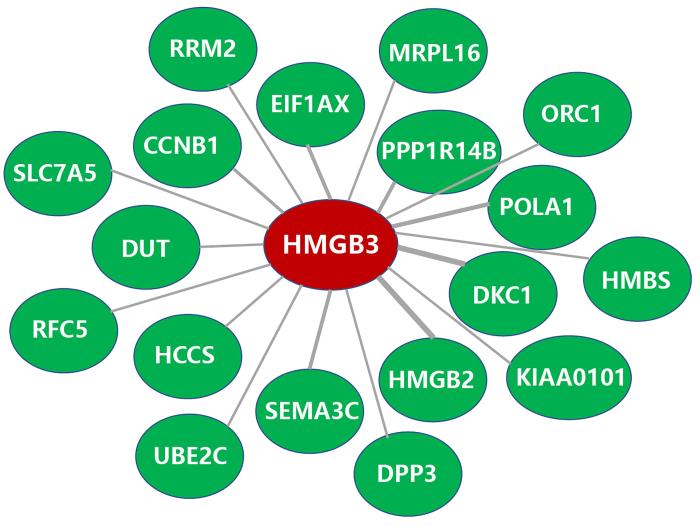
2.7 HMGB3的共表达网络与功能注释分析
通过Coexpedia获得与HMGB3共表达的分子调控网络,筛选出与乳腺癌相关的共表达分子共18个,包括HMGB2,DKC1,POLA1,PPP1R14B,EIF1AX,SEMA3C,CCNB1,RFC5,SLC7A5,DUT,KIAA0101,HCCS,RRM2,ORC1,MRPL16,HMBS,DPP3,UBE2C,如图5所示.
图5
图5
HMGB3在乳腺癌中的共表达分子网络
Fig.5
Network of HMGB3 co⁃expressed molecules in breast cancer
使用STRING数据库进行HMGB3和其18个共表达基因蛋白的功能注释分析.生物过程方面,九个基因蛋白在DNA代谢过程起重要作用,六个基因蛋白参与DNA复制.细胞成分方面,多个基因蛋白主要分布在染色体和核质中.分子功能方面主要与DNA结合弯曲和DNA聚合酶活性等相关.信号通路方面,主要参与调控P53信号通路、细胞周期信号通路和药物代谢等.如表2所示.
表2 HMGB3及其共表达基因的功能注释分析
Table 2
| subtype | functional annotation | FDR | genes |
|---|---|---|---|
| Biological process | DNA metabolic process | 1.50e-05 | HMGB2,KIAA0101,RRM2,DKC1, HMGB3,ORC1,POLA1,DUT,RFC5 |
| DNA replication | 1.56e-05 | KIAA0101,RRM2,ORC1,POLA1,DUT,RFC5 | |
| DNA biosynthetic process | 0.00071 | KIAA0101,DKC1,POLA1,RFC5 | |
| regulation of transcription involved in G1/S transition of mitotic cell cycle | 0.00089 | RRM2,ORC1,POLA1 | |
| mitotic cell cycle phase transition | 0.00089 | CCNB1,UBE2C,RRM2,ORC1,POLA1 | |
| cell cycle process | 0.0017 | CCNB1,KIAA0101,UBE2C,RRM2,ORC1,POLA1,RFC5 | |
| Cellular component | chromosome | 0.023 | CCNB1,HMGB2,HMGB3,ORC1,POLA1,RFC5 |
| nucleoplasm | 0.031 | CCNB1,HMGB2,KIAA0101,UBE2C, DPP3,DKC1,ORC1,POLA1,DUT,RFC5 | |
| nuclear chromosome | 0.0471 | CCNB1,HMGB2,ORC1,POLA1 | |
| replication fork | 0.0471 | POLA1,RFC5 | |
| Molecular function | four⁃way junction DNA binding | 0.009 | HMGB2, HMGB3 |
| DNA binding, bending | 0.0129 | HMGB2, HMGB3 | |
| DNA polymerase activity | 0.0178 | DUT, DKC1, EIF1AX, HMGB2, HMGB3 | |
| KEGG pathways | Pyrimidine metabolism | 0.0027 | RRM2,POLA1,DUT |
| Porphyrin and chlorophyll metabolism | 0.0064 | HMBS,HCCS | |
| DNA replication | 0.0064 | POLA1,RFC5 | |
| Drug metabolism:other enzymes | 0.0109 | RRM2,DUT | |
| p53 signaling pathway | 0.0109 | CCNB1,RRM2 | |
| Cell cycle | 0.0225 | CCNB1,ORC1 | |
| Purine metabolism | 0.0369 | RRM2,POLA1 |
FDR (False Discovery Rate),错误发现率,数值越小,相关基因与富集的功能关系越密切.
3 讨论
HMGB3是HMGB家族的重要成员之一,其编码的蛋白是一种非组蛋白染色质相关蛋白,在染色质的结构与功能及基因表达调控过程中均发挥着重要作用.非编码RNA可以调节HMGB3表达以促进包括胃癌[23]、肺癌[24-26]、鼻咽癌[27]、前列腺癌[28]、肝癌[29]、胶质母细胞瘤[30]等肿瘤细胞的增殖,侵袭和迁移并抑制癌细胞的凋亡.本研究通过TIMER数据库证实了HMGB3在包括乳腺癌在内的多种肿瘤中过表达.GEPIA和UALCAN数据库分析发现,乳腺癌中HMGB3转录水平和蛋白质表达较正常乳腺组织升高,而启动子甲基化水平较正常乳腺组织降低.由于其在乳腺癌中低甲基化,HMGB3可以被激活和上调,这提示HMGB3基因的甲基化修饰程度下调可能是乳腺癌中HMGB3表达增加的潜在机制.此外,通过PrognoScan数据库分析及Kaplan⁃Meier Plotter数据库分析表明HMGB3高表达预示乳腺癌不良预后.这些结果提示HMGB3可能在乳腺癌的发病过程中作为癌基因发挥作用,并且可以作为预后指标预测乳腺癌患者生存.
以往的研究表明HMGB3可以通过调节缺氧诱导因子1的表达来抑制肿瘤的增殖和乳腺癌的生长[31-32],HMGB3基因敲除实验[33]表明下调HMGB3表达可以抑制乳腺癌细胞的增殖和侵袭.本研究的亚组生存分析表明,HMGB3在ER阳性或luminal A型乳腺癌中预示患者更差的RFS和DMFS,因此,在ER阳性或luminal A型乳腺癌内分泌治疗耐药患者中靶向HMGB3可能是改善患者预后的潜在策略.而在淋巴结阳性状态下HMGB3高表达有更差的预后,可能提示HMGB3高表达与乳腺癌远处转移相关,监测乳腺癌HMGB3表达有助于预测乳腺癌远处转移,靶向HMGB3可能降低乳腺癌远处转移的发生.在TP53突变状态下HMGB3高表达患者有更差的预后,而TP53未突变状态下HMGB3高表达乳腺癌患者预后更好.沉默HMGB3可以调节P53和P21通路从而抑制细胞增殖和诱导胃癌细胞的G0/G1期停滞[34],功能分析表明HMGB3参与调节P53信号通路,提示HMGB3可能与TP53基因突变有关并参与调节P53信号通路促进乳腺癌发生发展并影响乳腺癌预后,但需要进一步实验加以验证.通过构建HMGB3基因的共表达分子网络,进行基因蛋白的功能注释分析显示,HMGB3与其共表达基因除了参与P53信号通路,还参与调控细胞周期信号通路和药物代谢.细胞周期信号通路参与调控细胞的生长、增殖及分化,通路中的许多因子起着原癌基因或抑癌基因的作用,这些因子的激活或畸变促进了癌症的发生发展[35].调节药物代谢酶是肿瘤细胞产生化学耐药性的机制之一[36-37],而HMGB3表达的上调可导致胃癌及卵巢癌发生化疗耐药[34,38].因此,HMGB3及其共表达基因可能通过调节这些通路在乳腺癌细胞增殖、侵袭、转移和耐药过程中发挥作用.以上结果表明HMGB3可作为乳腺癌的潜在治疗靶点,为进一步深入研究提供方向.
既往研究分析了TIL水平与乳腺癌亚型的关系,高TIL与非管腔亚型乳腺癌包括三阴性乳腺癌和HER2+乳腺癌更好地预后相关[39-40].分析不同种类TIL与乳腺癌预后的关系,结果表明,高水平的B细胞、CD8+T细胞、CD4+T细胞、中性粒细胞和树突状细胞浸润提示乳腺癌患者更好的10年累积生存,但巨噬细胞浸润水平与患者预后无关(图4).最新研究表明,组蛋白脱乙酰基酶3可通过下调miR⁃130a⁃3p来增加HMGB3表达,从而调节肿瘤免疫性T细胞水平以促进乳腺癌细胞的免疫逃逸[41].本研究中,高HMGB3表达在低浸润水平的B细胞、CD8+T细胞、CD4+T细胞、自然杀伤细胞和Th1细胞亚组中具有较差的预后(图5),提示HMGB3可能通过下调肿瘤中B细胞、CD8+T细胞、CD4+T细胞、自然杀伤细胞和Th1细胞浸润水平以促进乳腺癌细胞免疫逃逸从而影响乳腺癌患者预后.FOXP3是调节性T细胞的主要标志物,它会诱导免疫耐受从而干扰抗肿瘤免疫力,而乳腺癌患者中较高的FOXP3+TILs水平导致较差的总体生存率[42].发现富集的Treg细胞水平下HMGB3高表达乳腺癌患者预后更差,这提示HMGB3可能通过上调FOXP3+Treg细胞水平诱导肿瘤免疫耐受从而影响乳腺癌患者预后.还需进行进一步研究,以确认HMGB3表达在乳腺癌免疫微环境中的关键作用,并评估其表达与免疫细胞疗法的响应之间的关联.
本研究发现HMGB3在乳腺癌中高表达且与乳腺癌患者不良预后有关,可作为乳腺癌的预后指标和潜在治疗靶点.此外,HMGB3与乳腺癌免疫微环境的关系和功能分析结果为乳腺癌的靶向及免疫治疗研究提供了一个方向.但是由于数据库的限制,没有继续分析HMGB3与肿瘤免疫浸润的深层关系.
参考文献
Breast cancer incidence and mortality in women in China:Temporal trends and projections to 2030
Overexpression of high mobility group box 1 and 2 is associated with the progression and angiogenesis of human bladder carcinoma
Overexpression of high mobility group (HMG) B1 and B2 proteins directly correlates with the progression of squamous cell carcinoma in skin
Increased expression of HMGB3:A novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma
Overexpression of HMGB3 protein promotes cell proliferation,migration and is associated with poor prognosis in urinary bladder cancer patients
High mobility group box 3 as an emerging biomarker in diagnosis and prognosis of hepatocellular carcinoma
High⁃mobility group box 1 (HMGB1) promotes angiogenesis and tumor migration by regulating hypoxia⁃inducible factor 1 (HIF⁃1α) expression via the phosphatidylinositol 3⁃Kinase (PI3K)/AKT signaling pathway in breast cancer cells
HMGB1⁃activated fibroblasts promote breast cancer cells metastasis via RAGE/aerobic glycolysis
HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer
Mathematical modeling of the effect of boosting tumor infiltrating lymphocyte in immunotherapy
TIMER:A web server for comprehensive analysis of tumor⁃infiltrating immune cells
GEPIA:A web server for cancer and normal gene expression profiling and interactive analyses
UALCAN:A portal for facilitating tumor subgroup gene expression and survival analyses
PrognoScan:A new database for meta⁃analysis of the prognostic value of genes
Web⁃based survival analysis tool tailored for medical research (KMplot):Development and implementation
COEXPEDIA:Exploring biomedical hypotheses via co⁃expressions associated with medical subject headings (MeSH)
The STRING database in 2017:Quality⁃controlled protein⁃protein association networks,made broadly accessible
DNA methylation profiling in the clinic:Applications and challenges
DNA methylation and human disease
Aberrant CpG⁃island methylation has non⁃random and tumour⁃type⁃specific patterns
Tumor⁃infiltrating lymphocytes (TILs) in ER+/HER2⁃breast cancer
Prognostic role and clinical significance of tumor⁃infiltrating lymphocyte (TIL) and programmed death ligand 1 (PD⁃L1) expression in triple⁃negative breast cancer (TNBC):A systematic review and meta⁃analysis study
Upregulation of miR⁃513b inhibits cell proliferation,migration,and promotes apoptosis by targeting high mobility group⁃box 3 protein in gastric cancer
LncRNA PITPNA⁃AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA
Effects of microRNA⁃513b on cell proliferation,apoptosis,invasion,and migration by targeting HMGB3 through regulation of mTOR signaling pathway in non⁃small⁃cell lung cancer
Overexpression of miR⁃758 inhibited proliferation,migration,invasion,and promoted apoptosis of non⁃small cell lung cancer cells by negatively regulating HMGB
LncRNA SNHG5 promotes nasopharyngeal carcinoma progression by regulating miR⁃1179/HMGB3 axis
Long noncoding RNA SOX2⁃OT knockdown inhibits proliferation and metastasis of prostate cancer cells through modulating the miR⁃452⁃5p/HMGB3 axis and inactivating Wnt/β⁃catenin pathway
Upregulation of miR⁃200b inhibits hepatocellular carcinoma cell proliferation and migration by targeting HMGB3 protein
HMGB3 promotes the proliferation and metastasis of glioblastoma and is negatively regulated by miR⁃200b⁃3p and miR⁃200c⁃3p
HMGB3 small interfere RNA suppresses mammosphere formation of MDA⁃MB⁃231 cells by down⁃regulating expression of HIF1α
HMGB3 silence inhibits breast cancer cell proliferation and tumor growth by interacting with hypoxia⁃inducible factor 1α
Tumor suppressive function of mir⁃205 in breast cancer is linked to HMGB3 regulation
Knockdown of high mobility group⁃box 3 (HMGB3) expression inhibits proliferation,reduces migration,and affects chemosensitivity in gastric cancer cells
Perspective:Defects in cell cycle control and cancer
Drug⁃metabolizing enzymes:Role in drug resistance in cancer
Drug metabolizing enzymes⁃associated chemo resistance and strategies to overcome it
Targeting the high⁃mobility group box 3 protein sensitizes chemoresistant ovarian cancer cells to Cisplatin
Diffuse distribution of tumor⁃infiltrating lymphocytes is a marker for better prognosis and chemotherapeutic effect in triple⁃negative breast cancer
Predictive and prognostic role of tumour⁃infiltrating lymphocytes in breast cancer patients with different molecular subtypes:A meta⁃analysis
HDAC3 increases HMGB3 expression to facilitate the immune escape of breast cancer cells via down⁃regulating microRNA⁃130a⁃3p
Worse outcome in breast cancer with higher tumor⁃infiltrating FOXP3+Tregs:A systematic review and meta⁃analysis