南京大学学报(自然科学版) ›› 2020, Vol. 56 ›› Issue (3): 418–429.doi: 10.13232/j.cnki.jnju.2020.03.010
基因转录爆发的建模研究
- 南京大学物理学院,南京,210093
Modeling of transcriptional bursting
- School of Physics,Nanjing University,Nanjing,210093,China
摘要:
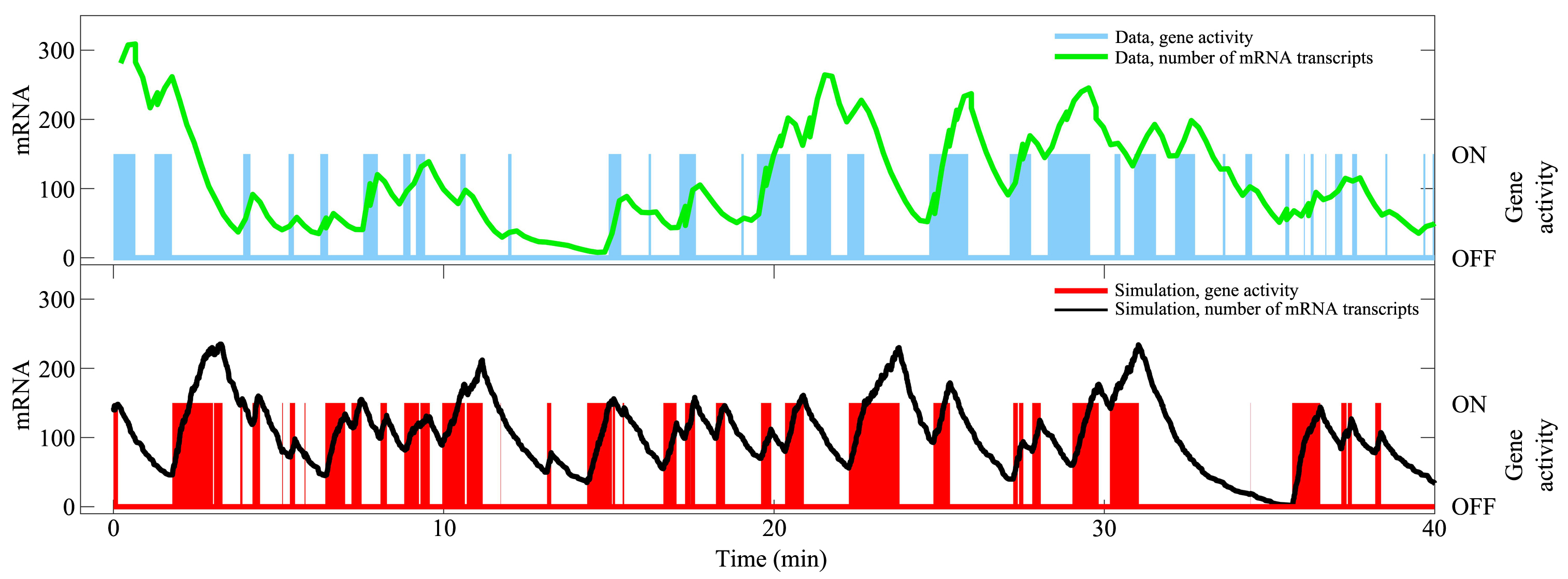
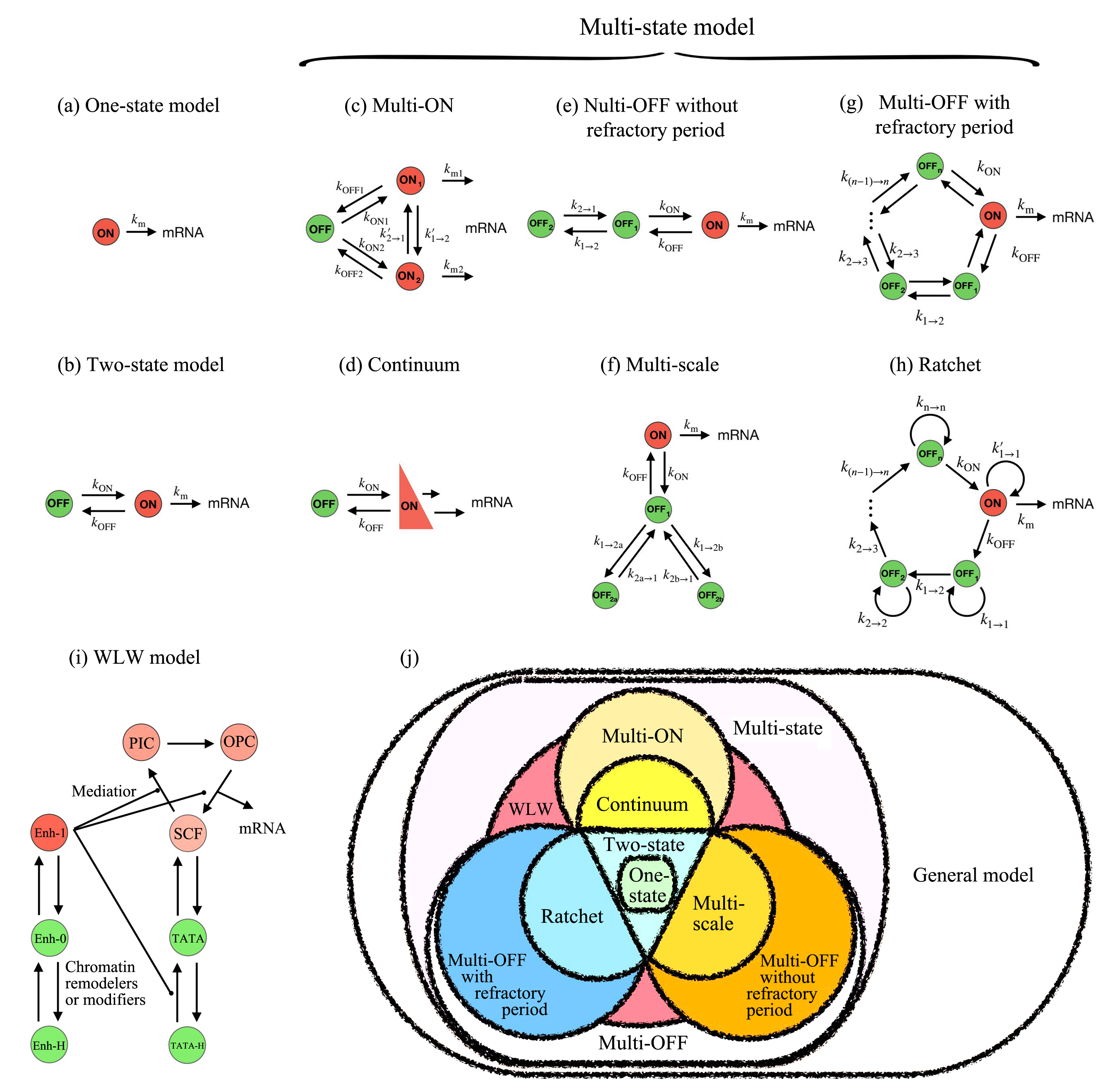
基因转录是细胞最重要的活动之一,涉及众多分子事件,且不同基因间存在显著差异性.建立基因转录的模型有助于理解复杂的转录动力学和调控机制.如何构建合适的转录模型依然具有大的挑战性.近年来的实验发现,转录爆发是一类普遍的转录模式,揭示其特征(如转录爆发的频率和大小以及激活态和沉默态的持续时间等)和调控机制是当前的研究热点.人们相继提出两态模型和多态模型来理解转录现象.有些模型不再是简单的唯象模型,而是考虑了转录的分子过程,能够深入研究转录的内在机理.结合最近的实验和理论研究,综述不同转录模型的特点、合理性及其适用范围,特别比较了各个模型的优缺点,有助于在研究中选取合适的转录模型.随着单细胞实验技术的发展,构建基因转录的定量模型将起到越来越重要的作用.
中图分类号:
- Q615
| 1 | Jonkers I,Kwak H,Lis J T. Genome?wide dynamics of Pol II elongation and its interplay with promoter proximal pausing,chromatin,and exons. eLife,2014,3:e02407. |
| 2 | Stasevich T J,Hayashi?Takanaka Y,Sato Y,et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature,2014,516(7530):272-275. |
| 3 | Senecal A,Munsky B,Proux F,et al. Transcription factors modulate c?Fos transcriptional bursts. Cell Reports,2014,8(1):75-83. |
| 4 | Voss T C,Hager G L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nature Reviews Genetics,2014,15(2):69-81. |
| 5 | Brown C R,Mao C,Falkovskaia E,et al. Linking stochastic fluctuations in chromatin structure and gene expression. PLoS Biology,2013,11(8):e1001621. |
| 6 | Nicolas D,Zoller B,Suter D M,et al. Modulation of transcriptional burst frequency by histone acetylation. Proceedings of the National Academy of Sciences of the United States of America,2018,115(27):7153-7158. |
| 7 | Muramoto T,Müller I,Thomas G,et al. Methylation of H3K4 is required for inheritance of active transcriptional states. Current Biology,2010,20(5):397-406. |
| 8 | Sanchez A,Choubey S,Kondev J. Stochastic models of transcription:from single molecules to single cells. Methods,2013,62(1):13-25. |
| 9 | Chubb J R,Trcek T,Shenoy S M,et al. Transcriptional pulsing of a developmental gene. Current Biology,2006,16(10):1018-1025. |
| 10 | Golding I,Paulsson J,Zawilski S M,et al. Real?time kinetics of gene activity in individual bacteria. Cell,2005,123(6):1025-1036. |
| 11 | Raj A,Peskin C S,Tranchina D,et al. Stochastic mRNA synthesis in mammalian cells. PLoS Biology,2006,4(10):e309. |
| 12 | Suter D M,Molina N,Gatfield D,et al. Mammalian genes are transcribed with widely different bursting kinetics. Science,2011,332(6028):472-474. |
| 13 | Wang Y,Ni T,Wang W,et al. Gene transcription in bursting:a unified mode for realizing accuracy and stochasticity. Biological Reviews,2019,94(1):248-258. |
| 14 | Ko M S H. Induction mechanism of a single gene molecule:Stochastic or deterministic? BioEssays,1992,14(5):341-346. |
| 15 | Zhang J,Zhou T. Promoter?mediated tran?scriptional dynamics. Biophysical Journal,2014,106(2):479-488. |
| 16 | Zhang J,Chen L,Zhou T. Analytical distribution and tunability of noise in a model of promoter progress. Biophysical Journal,2012,102(6):1247-1257. |
| 17 | Zhou T,Zhang J. Analytical results for a multistate gene model. SIAM Journal on Applied Mathematics,2012,72(3):789-818. |
| 18 | Bertrand E,Chartrand P,Schaefer M,et al. Localization of ASH1 mRNA particles in living yeast. Molecular Cell,1998,2(4):437-445. |
| 19 | Femino A M,Fay F S,Fogarty K,et al. Visualization of single RNA transcripts in situ. Science,1998,280(5363):585-590. |
| 20 | Zenklusen D,Larson D R,Singer R H. Single?RNA counting reveals alternative modes of gene expression in yeast. Nature Structural & Molecular Biology,2008,15(12):1263-1271. |
| 21 | Gillespie D T. Exact stochastic simulation of coupled chemical reactions. The Journal of Physical Chemistry,1977,81(25):2340-2361. |
| 22 | Pedraza J M,Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science,2008,319(5861):339-343. |
| 23 | Zoller B,Nicolas D,Molina N,et al. Structure of silent transcription intervals and noise characteristics of mammalian genes. Molecular Systems Biology,2015,11(7):823. |
| 24 | Sepúlveda L A,Xu H,Zhang J,et al. Measurement of gene regulation in individual cells reveals rapid switching between promoter states. Science,2016,351(6278):1218-1222. |
| 25 | Fritzsch C,Baumg?rtner S,Kuban M,et al. Estrogen-dependent control and cell?to?cell varia?bility of transcriptional bursting. Molecular Systems Biology,2018,14(2):e7678. |
| 26 | Zoller B,Little S C,Gregor T. Diverse spatial expression patterns emerge from unified kinetics of transcriptional bursting. Cell,2018,175(3):835-847.e5. |
| 27 | Ochab?Marcinek A,Tabaka M. Bimodal gene expression in noncooperative regulatory systems. Proceedings of the National Academy of Sciences of the United States of America,2010,107(51):22096-22101. |
| 28 | To T L,Maheshri N. Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science,2010,327(5969):1142-1145. |
| 29 | Raj A,van Oudenaarden A. Nature,nurture,or chance:stochastic gene expression and its consequences. Cell,2008,135(2):216-226. |
| 30 | Peccoud J,Ycart B. Markovian modeling of gene?product synthesis. Theoretical Population Biology,1995,48(2):222-234. |
| 31 | Dar R D,Razooky B S,Singh A,et al. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proceedings of the National Academy of Sciences of the United States of America,2012,109(43):17454-17459. |
| 32 | Rodriguez J,Ren G,Day C R,et al. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell,2018,176(1-2):213-226.e18. |
| 33 | Harper C V,Finkenst?dt B,Woodcock D J,et al. Dynamic analysis of stochastic transcription cycles. PLoS Biology,2011,9(4):e1000607. |
| 34 | Corrigan A M,Tunnacliffe E,Cannon D,et al. A continuum model of transcriptional bursting. eLife,2016,5:e13051. |
| 35 | Tantale K,Mueller F,Kozulic?Pirher A,et al. A single?molecule view of transcription reveals convoys of RNA polymerases and multi?scale bursting. Nature Communications,2016,7:12248. |
| 36 | Krasnov A N,Mazina M Y,Nikolenko J V,et al. On the way of revealing coactivator complexes cross?talk during transcriptional activation. Cell & Bioscience,2016,6:15. |
| 37 | Lemaire V,Lee C F,Lei J,et al. Sequential recruitment and combinatorial assembling of multiprotein complexes in transcriptional activation. Physical Review Letters,2006,96(19):198102. |
| 38 | Wang Y,Liu F,Li J,et al. Reconciling the concurrent fast and slow cycling of proteins on gene promoters. Journal of the Royal Society Interface,2014,11(96):20140253. |
| 39 | Wang Y,Liu F,Wang W. Dynamic mechanism for the transcription apparatus orchestrating reliable responses to activators. Scientific Reports,2012,2:422. |
| 40 | Kornberg R D. Mediator and the mechanism of transcriptional activation. Trends in Biochemical Sciences,2005,30(5):235-239. |
| 41 | Malik S,Roeder R G. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends in Biochemical Sciences,2005,30(5):256-263. |
| 42 | Huh D,Paulsson J. Random partitioning of molecules at cell division. Proceedings of the National Academy of Sciences of the United States of America,2011,108(36):15004-15009. |
| 43 | Peterson J R,Cole J A,Fei J,et al. Effects of DNA replication on mRNA noise. Proceedings of the National Academy of Sciences of the United States of America,2015,112(52):15886-15891. |
| 44 | Padovan?Merhar O,Nair G P,Biaesch A G,et al. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Molecular Cell,2015,58(2):339-352. |
| 45 | Chen J,Zhang Z,Li L,et al. Single?molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell,2014,156(6):1274-1285. |
| 46 | Paakinaho V,Presman D M,Ball D A,et al. Single?molecule analysis of steroid receptor and cofactor action in living cells. Nature Communications,2017,8:15896. |
| 47 | Grimm J B,English B P,Chen J J,et al. A general method to improve fluorophores for live?cell and single-molecule microscopy. Nature Methods,2015,12(3):244-250. |
| 48 | Phillips R,Belliveau N M,Chure G,et al. Figure 1 theory meets figure 2 experiments in the study of gene expression. Annual Reviews of Biophysics,2019,48:121-163. |
| No related articles found! |
|